Zinc finger proteins are natural human proteins
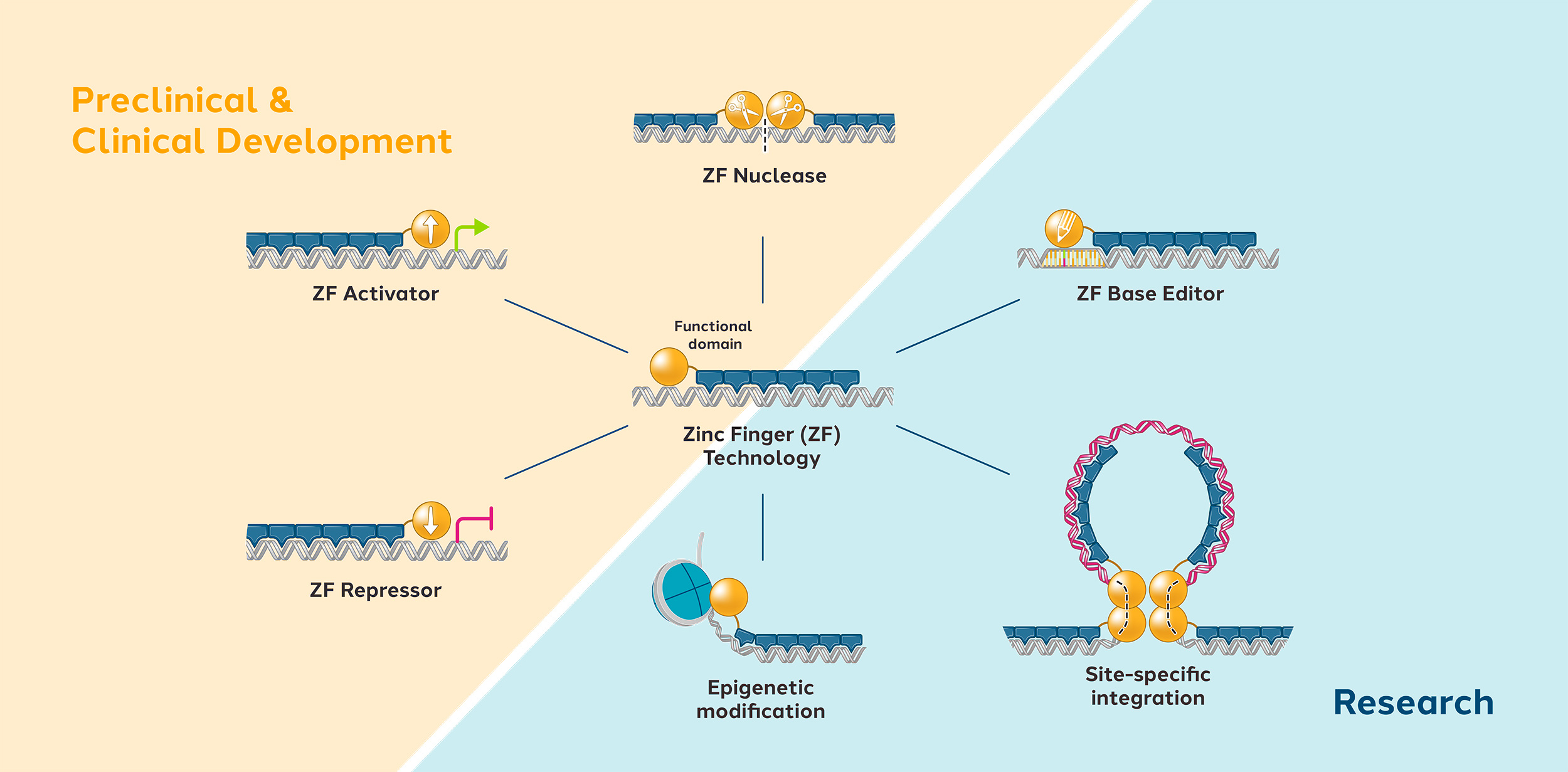
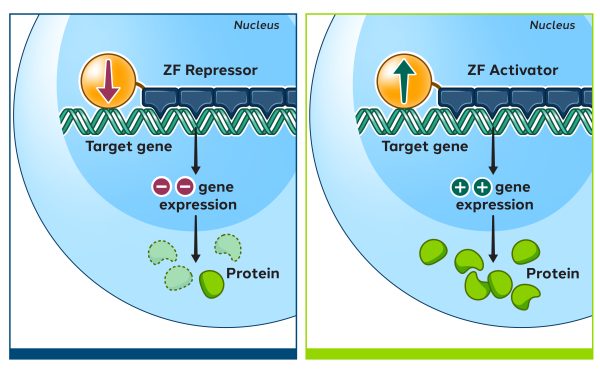
Zinc finger proteins (ZFPs) are naturally occurring proteins in humans which can recognize and bind to specific DNA sequences. Their natural function is to activate or repress the expression of human genes by turning them on or off. Using natural human proteins is expected to offer better safety compared to technologies derived from non-human sources.
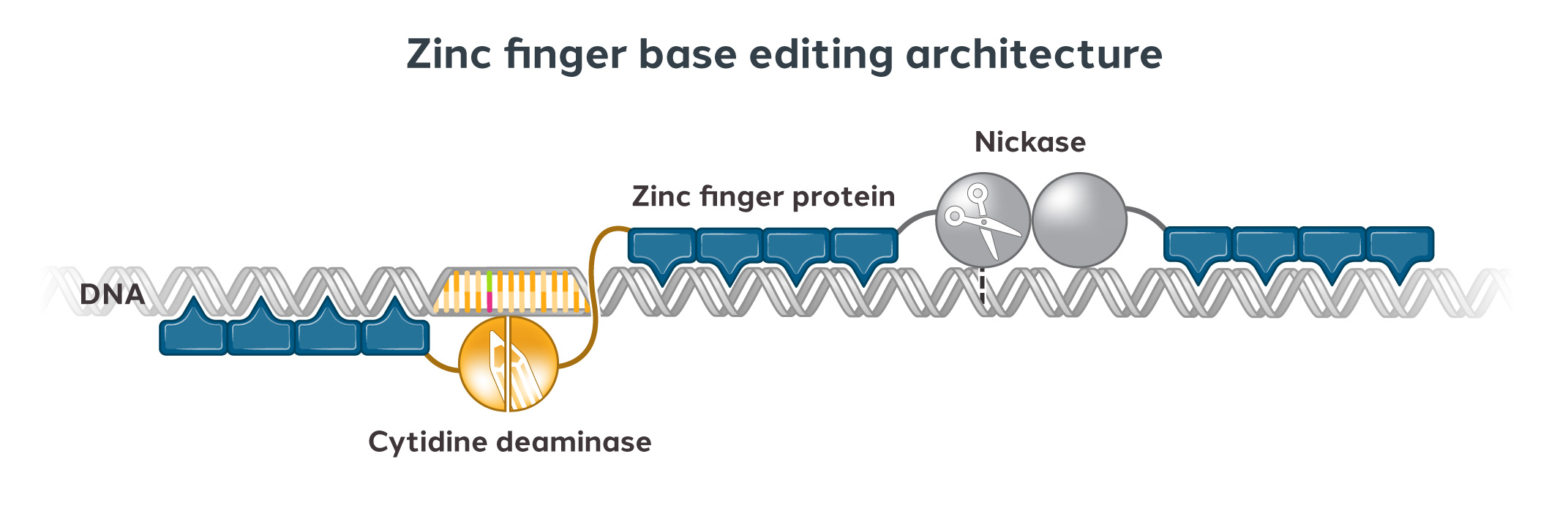
We can engineer ZFPs to recognize any unique DNA sequence of our choosing. Our modular approach usually involves using four to six zinc fingers, each of which can recognize a short DNA sequence. We can link multiple fingers together to form a zinc finger array that can recognize longer stretches of DNA, thus improving specificity.
We then attach the zinc finger array to a functional domain which imparts a change to the targeted DNA sequence, such as editing, repressing, or activating a gene.